Author: liuzlab
Using AI to improve diagnosis of rare genetic disorders
Diagnosing rare Mendelian disorders is a labor-intensive task, even for experienced geneticists. Investigators at Baylor College of Medicine are trying to make the process more efficient using artificial intelligence. The
Texas Children’s Hospital Inaugurates a New Data Science Center
Texas Children’s Hospital recently inaugurated a new Data Science Center on the first floor of the Jan and Dan Duncan Neurological Research Institute (Duncan NRI) that will serve as a
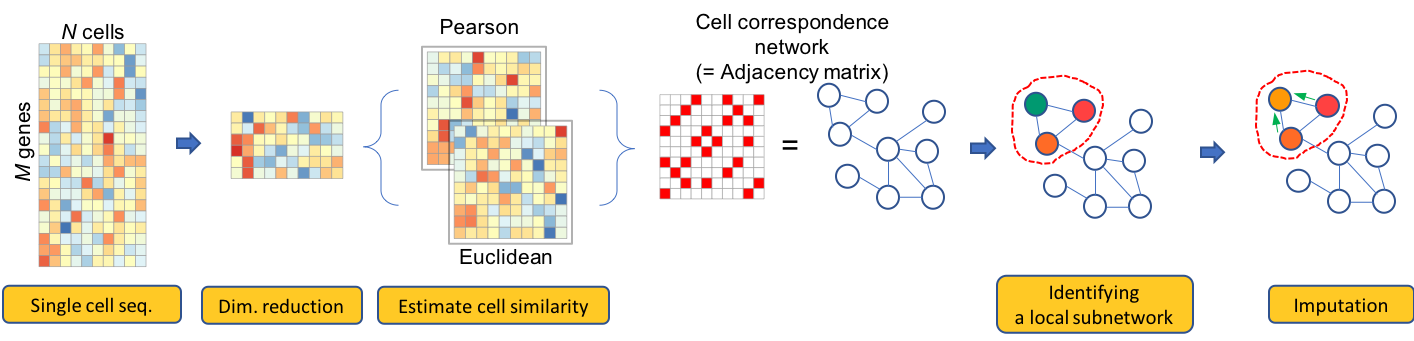
Our scRNA-seq imputation paper has been published in Bioinformatics.
Abstract Single-cell RNA sequencing technology provides a novel means to analyze the transcriptomic profiles of individual cells. The technique is vulnerable, however, to a type of noise called dropout effects,
We won the Allen Institute Cell Lineage Reconstruction DREAM Challenge
Jingyuan Hu and Dr. Zhandong Liu won the subchallenge 1 of the Allen Institute Cell Lineage Reconstruction DREAM Challenge! Congratulations! Details and list of the winners are available at: https://alleninstitute.org/what-we-do/frontiers-group/news-press/press-resources/press-releases/competition-yields-new-approaches-trace-development-cell-cell
Our analysis on the link between AD and HHV virus is published on Neuron.
https://www.sciencedaily.com/releases/2019/12/191218153350.htm
Dr. Hari Krishna Yalamanchili receives the 2018 NRI Zoghbi Scholar Award
Congratulations, Dr. Hari Krishna Yalamanchili for winning the NRI Zoghbi Scholar Award! He receives $50,000 to pursue innovative projects that will form the foundation of their independent research programs. Dr. Hari
Our long gene paper has been published in Nature Communications!
Our paper entitled “Apparent bias toward long gene misregulation in MeCP2 syndromes disappears after controlling for baseline variations” has been published in Nature Communications! Congratulations! Apparent bias toward long gene
We are hiring!
Bioinformatics Analyst/Programmer About the position: A bioinformatics analyst or programmer position is immediately available at the lab of Dr. Zhandong Liu from the Baylor College of Medicine at the Neurological
Dr. Liu receives distinguished service award!
Dr. Liu has received a distinguished service award from the International Association for Intelligent Biology and Medicine (IAIBM), congrats! Source: https://nri.texaschildrens.org/news/dr-zhandong-liu-receives-distinguished-service-award
Our transposable element paper has been published in Cell Reports!
Our paper entitled “TauActivates Transposable Elements in Alzheimer’s Disease” has been published in Cell Reports! Congrats! Tau Activates Transposable Elements in Alzheimer’s Disease Cell Reports, Volume 23, Issue 10, p2874–2880,
Congratulations to Hari and Aya for their Nature Neuroscience paper!
Hari’s paper that shows “Genome-wide distribution of linker histone H1.0 is independent of MeCP2.” is out now! Congrats Hari! Ito-Ishida A, Yamalanchili HK, Shao Y, Baker SA, Heckman LD, Lavery
Our Transposable Element quantification pipeline paper has been accepted by PSB 2018.
Our paper entitled “An ultra-fast and scalable quantification pipeline for transposable elements from next generation sequencing data” has been accepted by Pacific Symposium on Biocomputing (PSB) 2018, and the proceeding
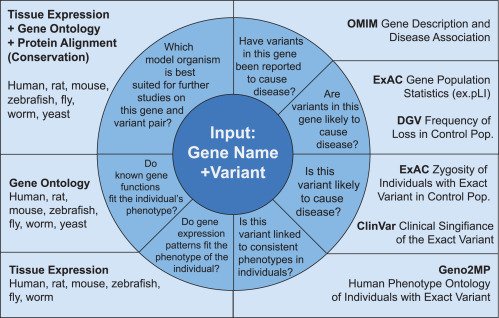
Our MARRVEL is selected as a Best of AJHG 2016-2017!
We are excited to announce that our MARRVEL paper is featured as one of the best papers of AJHG in late 2016 and early 2017! The AJHG journal selects best
Our paper on Batch Detection is out in Bioinformatics!
Ayush’s paper on batch detection is out now! Congrats Ayush! Detecting hidden batch factors through data adaptive adjustment for biological effects DASC, the batch detection tool is available in Bioconductor: https://bioconductor.org/packages/devel/bioc/html/DASC.html Abstract
Our CRISPRcloud paper has been published in Bioinformatics!
CRISPRcloud: A secure cloud-based pipeline for CRISPR pooled screen deconvolution Hyun-Hwan Jeong, Seon Young Kim, Maxime WC Rousseaux, Huda Y Zoghbi, Zhandong Liu We present a user-friendly, cloud-based, data analysis
Our MARRVEL paper has been published in AJHG!
MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome Julia Wang*, Rami Al-Ouran*, Yanhui Hu*, Seon-Young Kim*, Ying-Wooi Wan, Michael F. Wangler,
Our collaborative work is accepted by Nature Genetics.
Disruption of the ATXN1-CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans.
CPRIT Computational Award
Three grants totaling $2,634,668 were awarded to Individual Investigator Research Awards for Computational Biology. Our group is fortunate to be funded by CPRIT for our work on “Development and Validation of
Deep into mRNA-splicing – CrypSplice
Hari developed a new tool to discover non-canonical splicing events in RNA-seq: http://www.35.164.194.24/CrypSplice/
TCGA2STAT and protein data portal.
https://followthedata.wordpress.com/tag/r/
Congratulations to Wooi and Sztainberg for their Nature Paper.
Copy number variations have been frequently associated with developmental delay, intellectual disability and autism spectrum disorders. MECP2 duplication syndrome is one of the most common genomic rearrangements in males and
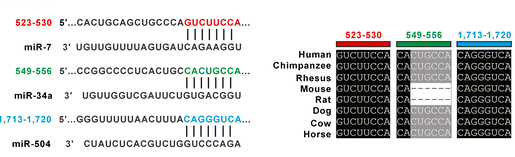
MicroRNA and Shank3 work published in molecular brain.
Post-transcriptional regulation of SHANK3 expression by microRNAs related to multiple neuropsychiatric disorders. Our study provides new insight into the miRNA-mediated regulation of SHANK3 expression, and its potential implication in multiple neuropsychiatric disorders
Our new profile on Benefunder.
http://benefunder.org/causes/549/zhandong-liu
The Siemens Competition in Math, Science & Technology
Congratulations to Kanchana to be selected as the semis-finalist of The Siemens Competition in Math, Science & Technology. Great job.
Congrats to Kanchana Raja for getting a first place award from the American Statistical Society and third place award in the Computer Science division.
Congratulations to Kanchana Raja for winning a first place award from the American Statistical Society and third place award in the Computer Science division in Houston Science Fair. Kanchana a
Congrats to Andrew Laitman for getting the NLM training fellowship!!!
Congrats to Andrew Laitman for getting the NLM training fellowship!!!
Joint work with Dr. Anderson funded by CPRIT.
Our project on Viral MicroRNAs in Ovarian Cancer Growth and Metastasis is funded through CPRIT.