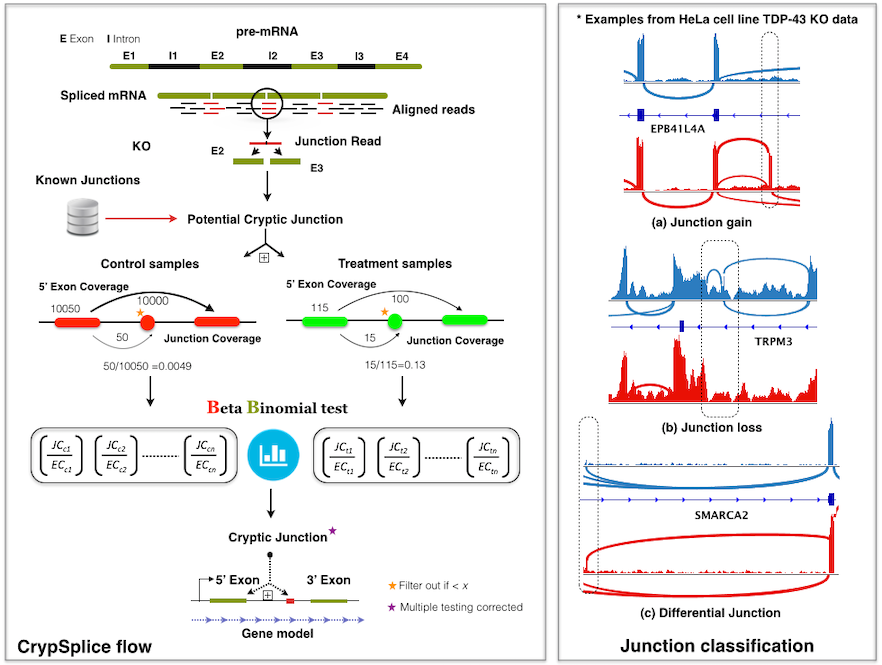
Alternative splicing of RNA is the key mechanism by which a single gene codes for multiple functionally diverse proteins. Recent studies identified previously unknown class of exons, ‘cryptic’ exons, in RNA transcripts. These cryptic exons are often associated with various human cancers and neurological disorders. Genome-wise detection of cryptic splice sites can facilitate a comprehensive understanding of the underlying disease mechanisms and develop strategies that hope to resolve cryptic splicing with the ultimate goal of therapeutic applications. CrypSplic is a novel cryptic splice site detection method. It uses beta-binomial distribution to model junction count data. Every junction is subjected to a beta binomial test w.r.t conditions and classified to aid molecular inferences.
© 2015 Huda Zoghbi Lab and Zhandong Liu Lab, Jan and Dan Duncan Neurological Research Institute at Texas Children’s Hospital1250 Moursund St., Houston, TX
Availability
Please click here to download CrypSplice.
Under the terms of the GNU General Public License as published by the Free Software Foundation (version 3 or later) and No Warranty.
Command
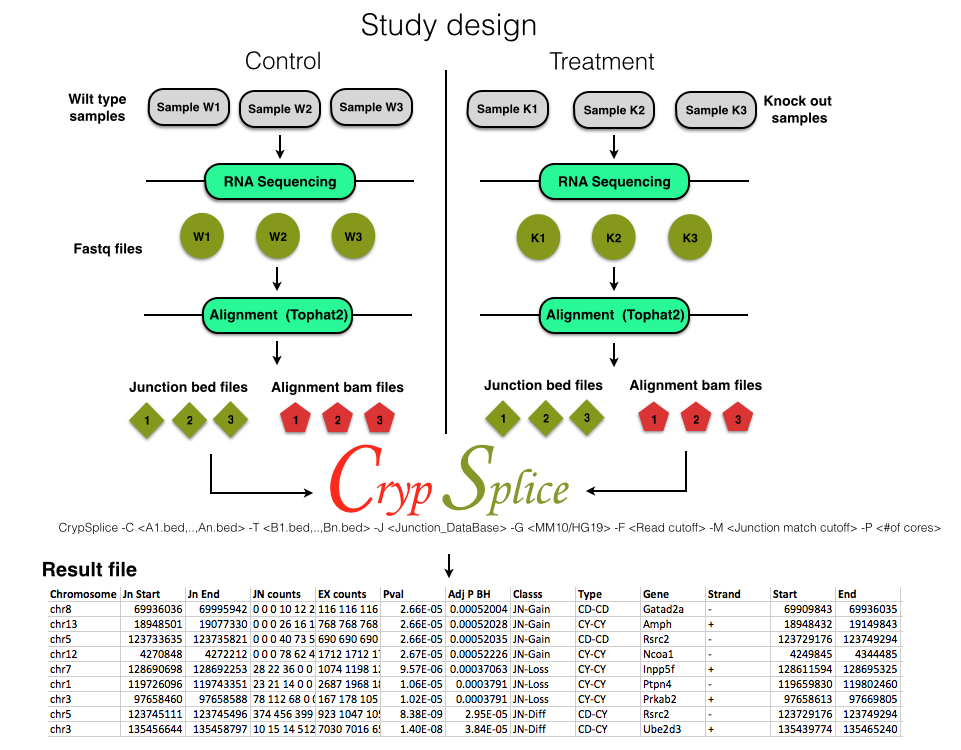
CrypSplice -C <A1.bed,..,An.bed> -T <B1.bed,..,Bn.bed> -G <MM10/HG19> -F <Read cutoff> -M <Junction match cutoff> -P <#of cores>
Input
CrypSplice accepts junction quantifications in bed format
chr1 990 2099 JUNC00001560 872 + 990 2099 255,0,0 2 40,67 0,1042
chr2 101 2121 JUNC00001561 16 + 1010 2121 255,0,0 2 20,63 0,1048
-C Junction bed files of control samples
-T Junction bed files of treatment samples
Tip: If junction files are not in BED format consider using regtools to extract junctions in BED format.
Parameters
-J Known Junctions data base in bed format.
-G Genome model directory containing respective alternativ splicing, gene model and Junction database files. Data for DM3, MM9, MM10, HG19, HG38 are provided. For other genome builds please use this MakeGeneModel script to generate the annotation model files.
Genome annotation related files including junction database (known junctions), known alternative splicing events and gene coordinates can be found in respective genome directories. If you are working with MM10 build: MM10-Known-SpliceJunctions.txt –> Junction database (Known junctions), MM10-Known-AE.txt –> Known splicing events and MM10.genes.bed —> gene coordinates.
-F Minimum expression cutoff. Junctions with coverage less than this are ignored. Default 10.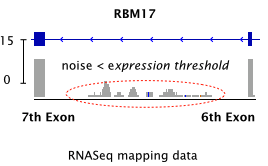
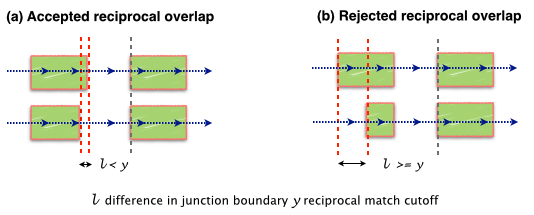
-M Juntion match cutoff. To account sequencing errors junctions with reciprocal overlap of M or more are collapsed. Default 0.95 (95%).
-P Number of CPU cores to use. Default 1.
Dependency
Bedtools V2.27 or higher to be accessed from cmd line
R libraries ibb and MASS
Note
CrypSplice also accommodates single sample data sets. However, we recommend users to use junction strength as a ranking metric.
To avoid confusion with multiple input files, we made -G parameter to handle all annotation related files. So if you provide -G you can ignore -J from earlier version.
For any queries please contact
Hari Krishna Y: hari.yalamanchili@bcm.edu
Zhandong Liu : zhandong.liu@bcm.edu